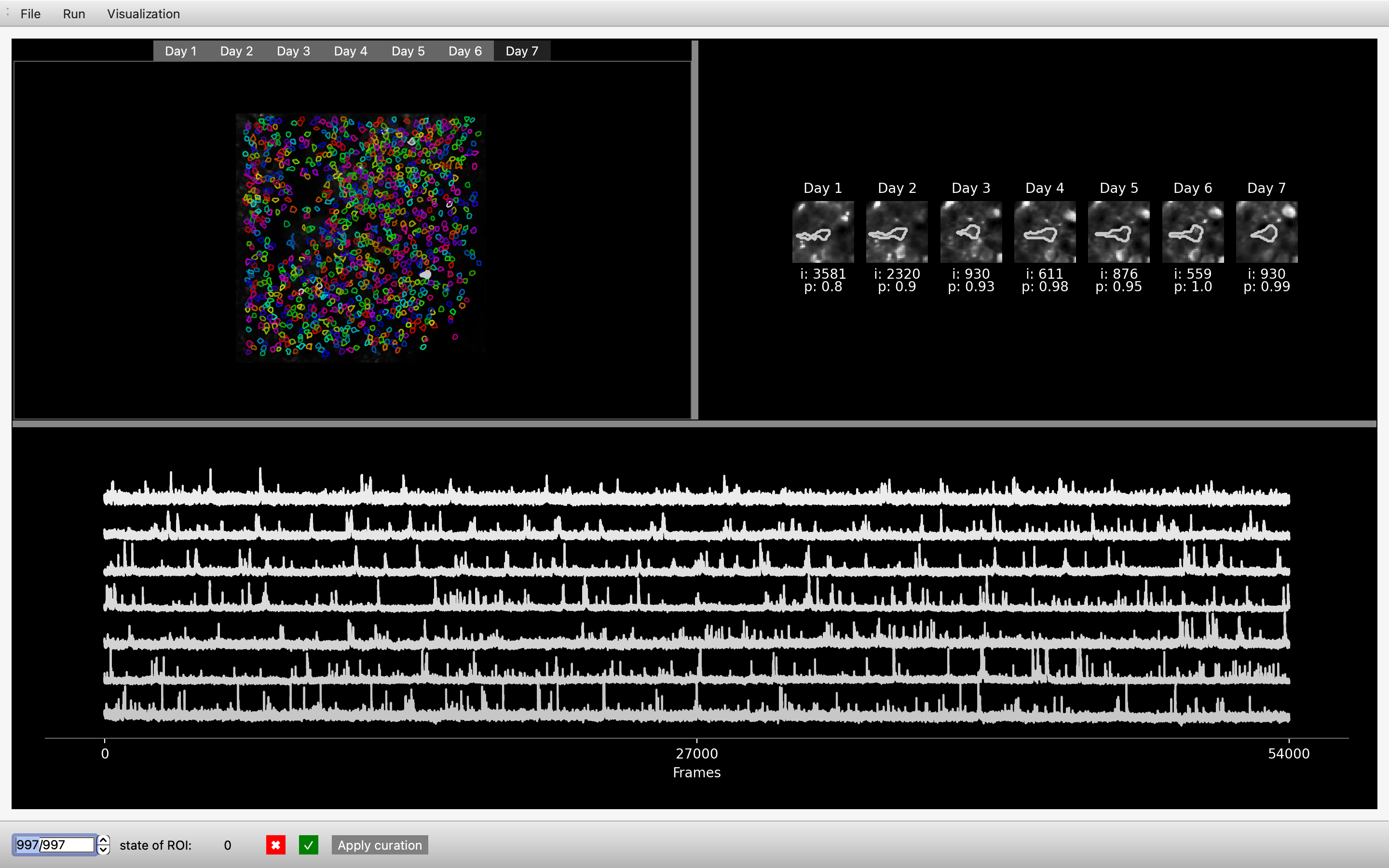
Curation of track2p outputs#
The bottom bar is dedicated to manual curation of track2p outpts. It is divided into several parts:
A text box with an up-down control: the user can browse all matches detected by the algorithm as present across all days, and all central window information will be updated according to the cell selected.
The interface allows the user to curate the results of the algorithm’s multi-day cell tracking. On initialization, all cells detected as being correctly tracked each day by our algorithm have a status of 1. This information is stored in track_ops.vector_curation (see parameters)
However, the user can assess the quality of cell tracking and activity by inspecting ROIs, zoomed images and the fluorescence traces across days. Subsequently, adjustments can be made to the status of a cell within track_ops.vector_curation through the interface.
State of ROI : informs the user of the current cell status stored in
track_ops.npy✅ : is used to set the cell state to 1 in
track_ops.vector_curation❌ : is used to set the cell state to 0 in
track_ops.vector_curationApply curation : is used to apply the curation to the GUI. When the user clicks on this button, all cells with a status of 0 will be white, while those with a status of 1 will be colored. Note: This button only affects the GUI visualisation. It is not necessary to click this button to save the curation to
track_ops.vector_curation, this is done automatically when the status of the cell is changed by the user.
Below is an example of a cell labelled as ‘not cell’ by the user (recording of developing mouse barrel cortex for consecutive 7 days)